mRNA’s importance
mRNA (messenger RNA) plays a crucial role in the flow of genetic information within a cell. mRNA acts as a messenger that carries genetic instructions from the DNA in the cell’s nucleus to the ribosomes in the cytoplasm, where proteins are synthesized. From the central dogma, which describes genetic information from DNA to RNA and then to proteins, we have learned the importance of RNA in last story.
Let’s delve deeper into RNA itself. Until now, three types of RNA have been considered crucial: messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA). Among these, mRNA contains the direct instructions for coding proteins. Therefore, textbooks primarily focus on these three types of RNA. Most people are familiar with the fact that mRNA uses codons—three-base sequences—between the start codon and stop codon to specify individual amino acids. Beyond this, extensive research has been conducted on mRNA, including mechanisms like the poly-A tail, which protects mRNA, and the 5′ cap, where RNA synthesis begins. Here, mRNA is an incredibly useful molecule in biology. By measuring the concentration of mRNA in a cell, we can infer the amount of protein present, as mRNA is absolutely essential for protein synthesis. mRNA analysis is typically performed using qPCR (quantitative PCR), which allows for precise measurement of mRNA levels in cells.
What is coding sequence (CDS)
The region of RNA that encodes the information for a protein, spanning from the start codon to the stop codon, is referred to as the Coding Sequence (CDS). Although RNA molecules that code for a single protein may vary in other regions (e.g., untranslated regions or poly-A tails), they ultimately produce the same protein if their CDS is identical. Therefore, when performing qPCR, the CDS should be the primary target to ensure the accurate quantification of the RNA related to the specific protein.
On the site, you input nucleotide and then specify the protein of interest.
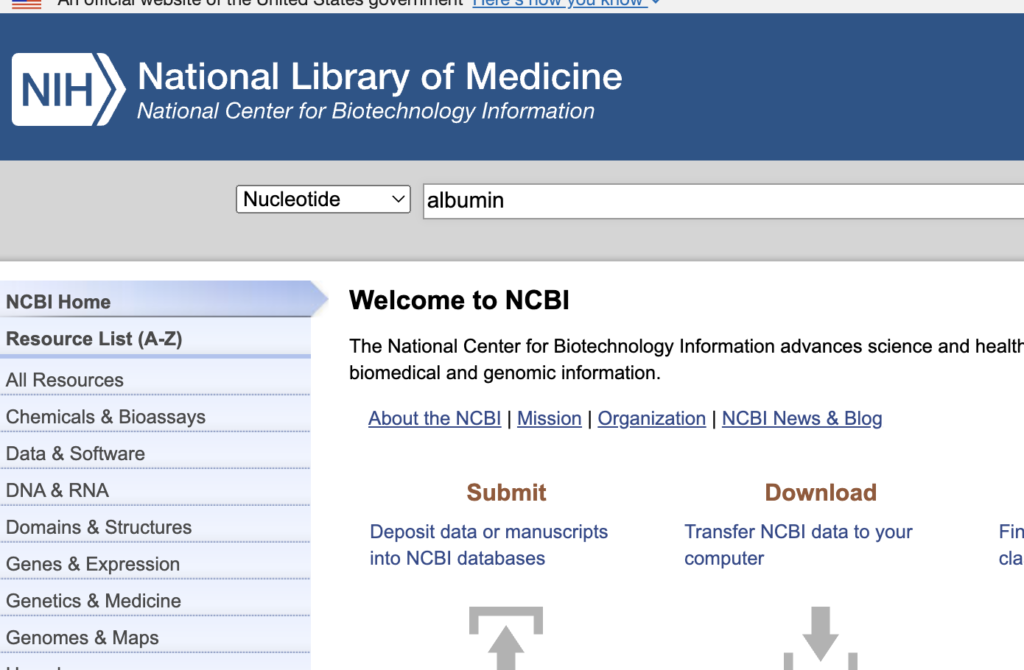
Click the gene name and select the species (Homo sapiens).
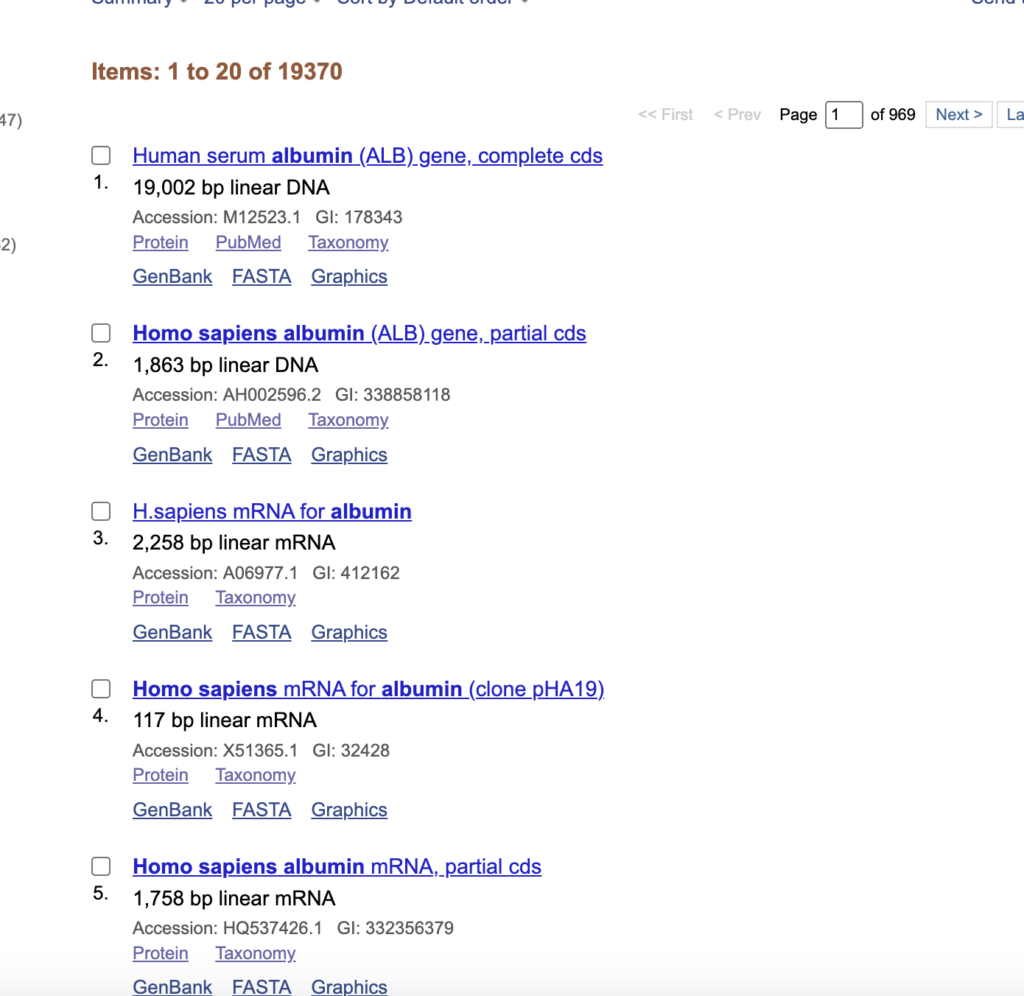
After select mRNA sequence (H.Sapiens mRNA for albumin)
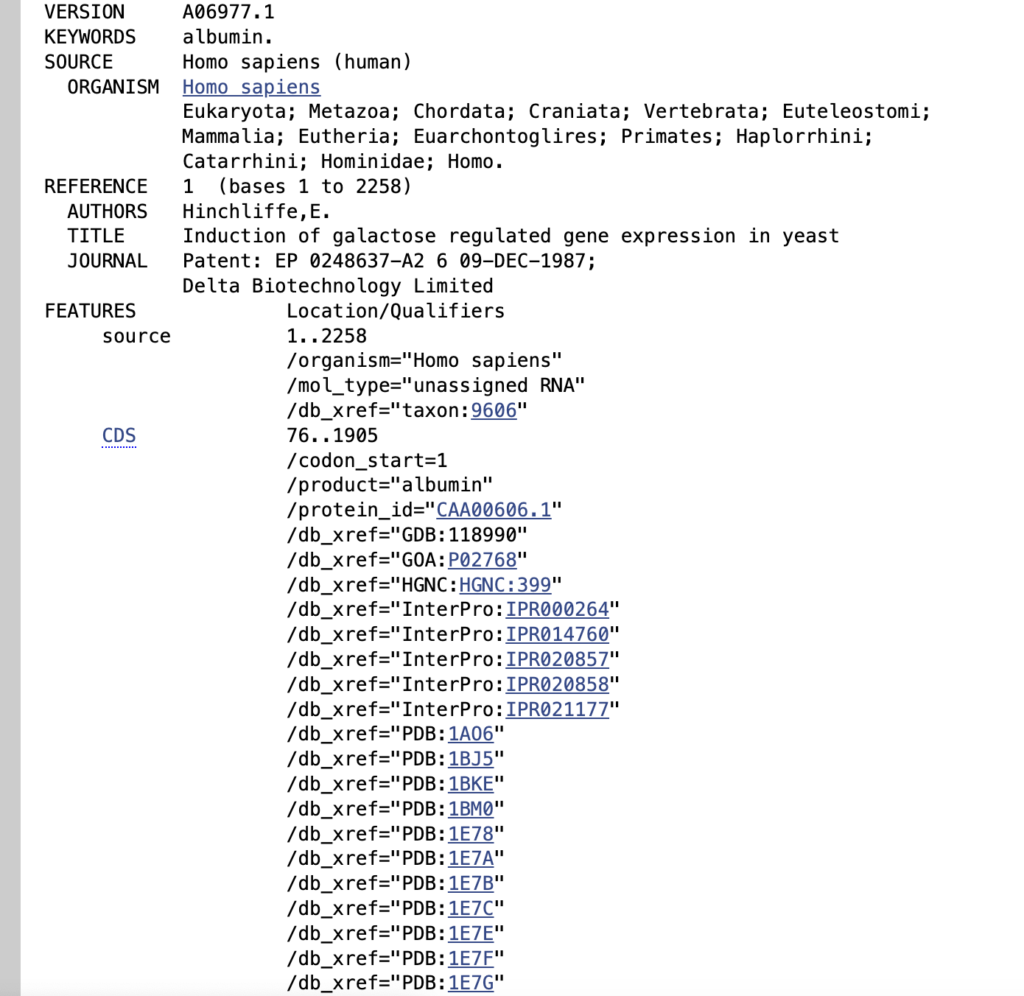
Click CDS (Coding Sequence)

You can locate the CDS sequence by identifying the ATG start codon and the TAA stop codon. The region between these two codons is the CDS (Coding Sequence). Any part of the sequence outside of these codons is considered the non-CDS part, such as untranslated regions (UTRs). If you want to find primer, you can use CDS region to Primer blast.
https://www.ncbi.nlm.nih.gov/tools/primer-blast
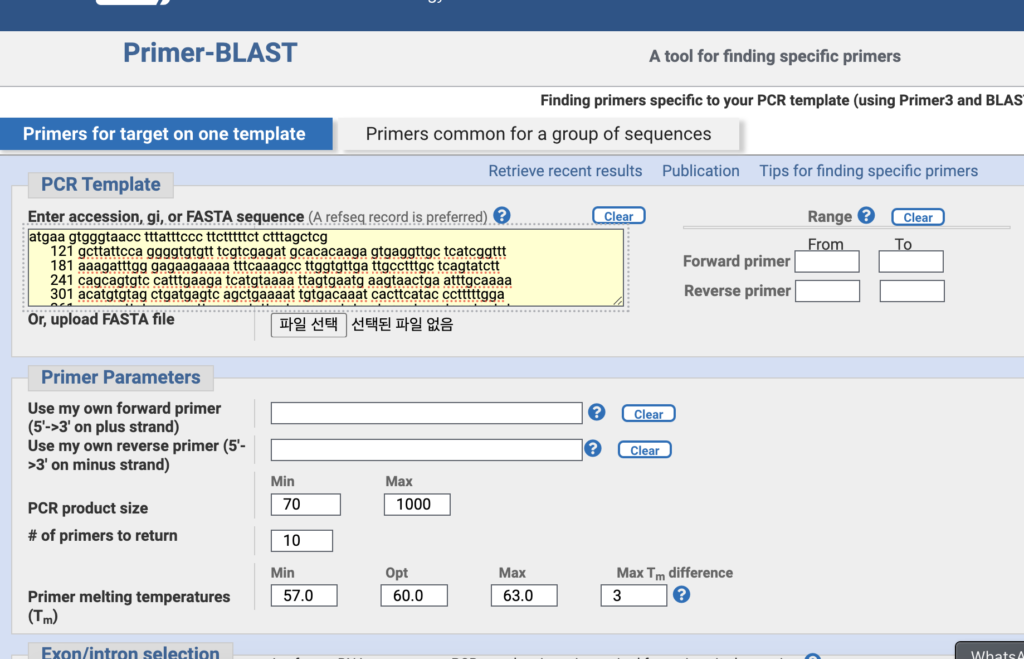
After copying the CDS sequence, paste it into the yellow section of the Primer BLAST tool. Adjust the other parameters accordingly. Generally, you prefer a PCR product size of about 90-150 bp, a Tm of 60°C, and a GC content between 45-55%. This will help you design primers that work well for your experiment.
Once the results are generated, you should select primers that show only a single mRNA for a specific protein. If more than one result appears, it means the result is based on multiple mRNA sequences, which reduces the reliability. Afterward, you need to check for self-loops and assess how well the two primers bind to each other. This will help ensure that the primers are designed correctly for your PCR experiment.
mRNA is a single-stranded molecule and is highly prone to degradation. Therefore, mRNA undergoes a process to convert it into a stable DNA form. This process results in the creation of cDNA (complementary DNA). cDNA is synthesized from RNA using a specific enzyme found in certain viruses, and this enzyme is called reverse transcriptase. This synthesis process is provided by manufacturers, so you can simply follow the manufacturer’s protocol, which is usually very straightforward and easy to perform.
cDNA and qPCR
For example, a product from the Japanese company Takara is commonly used for this process. Takara offers various kits for cDNA synthesis, and their protocols are well-established and easy to follow, making it a popular choice for researchers.
The cDNA generated is combined with the primers you designed earlier, and PCR is performed using SYBR Green to measure the amount of PCR product produced. The Ct (Cycle threshold) value is calculated based on the cycle at which the fluorescence signal rises sharply, indicating the presence of the PCR product. To quantify the gene expression, the Ct value of your target gene is compared to the Ct value of a housekeeping gene, such as GAPDH or beta-actin. This allows you to calculate the relative expression of the target gene by comparing the differences in Ct values. Based on this, you can compare the experimental group with the control group to determine any changes in mRNA expression. By examining the differences in the Ct values between the two groups, you can assess whether the target gene’s expression is upregulated or downregulated in response to the experimental conditions.
Even if the mRNA shows a high value, it is recommended to also examine protein-level expression if the gene is truly important. This is because mRNA is easily degraded and can be regulated by various factors, which means its expression might not always correlate with protein expression. By comparing both mRNA and protein expression, you can strongly support your desired hypothesis and provide more reliable evidence for your conclusions.

2 Replies to “mRNA and qPCR: Understanding Gene Expression (3)”